A team of researchers has developed an innovative method to design complicated all-α proteins, characterized by their non-uniformly arranged α-helices as seen in hemoglobin. Employing their novel approach, the team successfully created five unique all-α protein structures, each distinguished by their complicated arrangements of α-helices. This capability holds immense potential in designing functional proteins.
This research has been published in the journal “Nature Structural and Molecular Biology” on January 4, 2023.
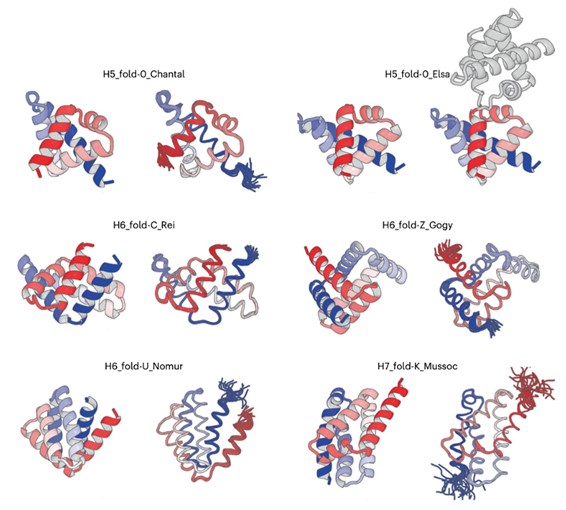
Figure: De novo designed all-α proteins with complicated shapes. H5_fold-0_Chantal, H5_fold-0_Elsa, H6_fold-C_Rei, H6_fold-Z_Gogy, H6_fold-U_Nomur, and H7_fold-K_Mussoc are presented. For each de novo designed protein, the computational model is shown on the left, and the solved experimental structure is on the right.
Figure Credit: Koya Sakuma, Naohiro Kobayashi, Toshihiko Sugiki, Toshio Nagashima, Toshimichi Fujiwara, Kano Suzuki, Naoya Kobayashi, Takeshi Murata, Takahiro Kosugi, Rie Tatsumi-Koga & Nobuyasu Koga, “Design of complicated all-α protein structures”, Nature Structural and Molecular Biology (2024)
Proteins fold into unique three-dimensional structures based on their amino acid sequences, which then dictate their function. Although significant progress has been made in de novo protein design, the ability to design complicated all-α proteins, where α-helices are non-parallelly arranged within the three-dimensional structures, was lacking. “Artificially designed proteins mostly show simple structures, but Nature presents us with complicated ‘designs’,” said Prof. Nobuyasu Koga of Exploratory Research Center on Life and Living Systems (ExCELLS) at National Institutes of Natural Sciences (NINS). This gap drove the team to seek a method to design such complicated all-α proteins.
The team began by examining structures deposited in the Protein Data Bank (PDB) and identified 18 typical helix-loop-helix motifs. They then demonstrated that a broad spectrum of all-α protein tertiary structures, ranging from simple to complicated, can be computationally generated by combining these identified typical motifs and canonical α-helices. “It is surprising that such a diverse set of all-α protein structures can be generated simply by combining typical or canonical components of naturally occurring proteins,” said Dr. Koya Sakuma, a former Ph. D. student in SOKENDAI (The Graduate University for Advanced Studies).
The team selected five unique all-α proteins, each with five or six α-helices and distinguished by their complicated spatial arrangements, from the computationally generated structures. They then carried out de novo design of amino acid sequences that are capable of folding into the selected five all-α protein structures.
The experimental results were remarkable (See the figure). “The structures that emerged from structure determination process exhibit the complicated shapes, which perfectly matched computationally designed structures,” said Dr. Naohiro Kobayashi, a senior research fellow at RIKEN.
With this developed method, it is now possible to create all-α proteins with complicated shapes. Functions of proteins inherently depend on their three-dimensional structures. If we can design proteins with complicated shapes, it increases the potential to construct functional sites within them. This breakthrough research will pave the way for the creation of new functional proteins, which will contribute to healthcare and life sciences.
The research team includes Koya Sakuma from SOKENDAI (currently, Nagoya University), Naohiro Kobayashi from RIKEN, Toshihiko Sugiki from the Institute for Protein Research (IPR), Osaka University (currently Kitasato University), Toshio Nagashima from RIKEN, Toshimichi Fujiwara from IPR, Osaka University, Kano Suzuki from Chiba University, Naoya Kobayashi from ExCELLS NINS (currently, Nara Institute of Science and Technology), Takeshi Murata from Chiba University, Takahiro Kosugi from Institute for Molecular Science (IMS) NINS, Rie Tatsumi-Koga from ExCELLS NINS (currently IPR, Osaka University), Nobuyasu Koga from IMS NINS, ExCELLS NINS, SOKENDAI (currently IPR, Osaka University).
FUNDER
Japan Agency for Medical Research and Development
the Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) program,
JJP19am0101072, JP20am0101083
Japan Science and Technology Agency (JST)
Precursory Research for Embryonic Science and Technology
JPMJPR13AD to N. Koga
Japan Society for the Promotion of Science(JSPS)
KAKENHI Grants-in-Aid for Scientific Research
15H05592 to N. Koga
18H05420 to T. Kosugi, N. Koga
18H05425 to T. Murata
18K06152 to Naohiro Kobayashi
JST-Mirai Program
JPMJMI17A2 to Naohiro Kobayashi
Grant-in-Aid for JSPS Fellows
15J02427 to K. Sakuma
Research Center for Computational Science, Okazaki
21-IMS-C174, 20-IMS-C157, 19-IMS-C175, 18-IMS-C155, 17-IMS-C147, 16-IMS-C129, 15-IMS-C180
Reference:
Authors: Koya Sakuma†, Naohiro Kobayashi†, Toshihiko Sugiki, Toshio
Nagashima, Toshimichi Fujiwara, Kano Suzuki, Naoya Kobayashi, Takeshi
Murata, Takahiro Kosugi, Rie Tatsumi-Koga, and Nobuyasu Koga*
(†co-first author、*corresponding author)
Journal Name: Design of complicated all-α protein structures
Journal Title: Nature Structural & Molecular Biology
DOI: 10.1038/s41594-023-01147-9
URL: https://www.nature.com/articles/s41594-023-01147-9
Expert Contact:
Institute for Protein Research (IPR), Osaka University
Professor Nobuyasu Koga
nkoga@protein.osaka-u.ac.jp
Media Contact:
Public Relations Manager
Institute for Molecular Science
press@ims.ac.jp
Public Relations Office
Chiba University
koho-press@chiba-u.jp
Recommend
-

Viewing a Diverse World through the Lens of Agriculture and Food: Enriching Global SDGs Education with Insights from Extensive Overseas Field Research
2023.08.31
-

The joy of revealing the unknown: The challenge of solving the mystery of the Sun
2022.09.28
-

The Science Behind Nature’s Touch: Horticultural Therapy and Community Well-Being
2025.01.14


