Researchers uncover how different mixing methods influence the molecular structure and gene-silencing efficiency of siRNA-loaded nanoparticles.
siRNA therapies show promise for treating diseases like cancer and genetic disorders, but their effectiveness depends on proper delivery. A recent study found that the method of mixing siRNA with lipid nanoparticles (LNPs) is key to success. Using NMR and small-angle X-ray scattering (SAXS), researchers revealed that different preparation methods affect the internal structure and siRNA distribution in LNPs, impacting their therapeutic potential. Optimizing these methods can enhance the efficacy of siRNA-loaded LNPs.
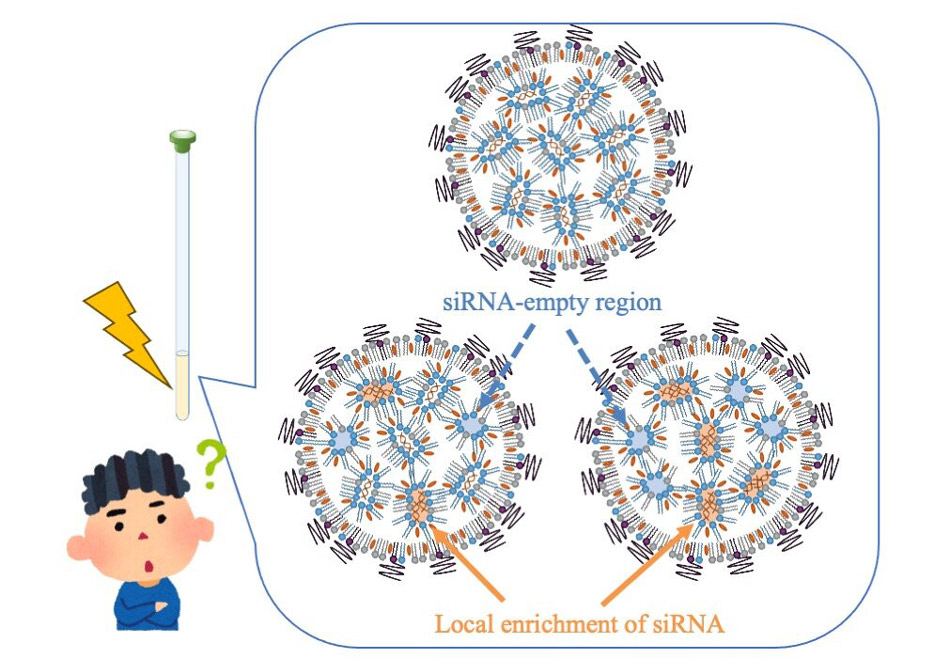
Image title: Molecular Heterogeneity in siRNA-Loaded LNPs
Image caption: Diagram of the image of being able to distinguish differences in the state of nucleic acids inside LNPs by evaluating their physical properties using NMR.
Image credit: Dr. Keisuke Ueda from Chiba University, Japan
Image license: Original content
Usage restrictions: Can be reused with proper attribution.
Small interfering RNA (siRNA) molecules hold immense potential for treating diseases by silencing specific genes. Encapsulated in lipid nanoparticles (LNPs), siRNA can be delivered efficiently to target cells. However, the effectiveness of these therapies hinges on the internal structure of the LNPs, which can significantly impact their ability to deliver siRNA. Traditional methods often fall short in providing the detailed molecular insights needed to fine-tune LNP design for optimal therapeutic efficacy.
A study published in the Journal of Controlled Release on August 02, 2024, led by Assistant Professor Keisuke Ueda from the Graduate School of Pharmaceutical Sciences, Chiba University, introduces a novel approach to improving siRNA-loaded LNPs. By employing NMR-based molecular-level characterization, the research investigates how different siRNA mixing methods affect the uniformity and molecular state of siRNA within LNPs. This research was coauthored by Dr. Hidetaka Akita from the Graduate School of Pharmaceutical Sciences, Tohoku University; Dr. Kenjirou Higashi from the Graduate School of Pharmaceutical Sciences, Chiba University; and Dr. Kunikazu Moribe (last author) from the Graduate School of Pharmaceutical Sciences, Chiba University.
“NMR allowed us to peer inside these nanoparticles at a molecular level, revealing the intricate details of how siRNA is distributed within the LNP core. This level of insight is crucial for understanding and optimizing LNP formulations,” said Dr. Ueda.
The team compared three preparation methods for siRNA-loaded LNPs to understand their impact on molecular structure and gene-silencing efficiency. The methods included pre-mixing, where siRNA and lipids were combined using a microfluidic mixer; post-mixing (A), where siRNA was mixed with empty LNPs in an acidic condition with ethanol; and post-mixing (B), where siRNA was mixed with empty LNPs in an acidic condition without ethanol.
While all three methods produced LNPs with a consistent size of about 50nm and maintained a constant ratio of siRNA to lipid content, the distribution of siRNA within the LNPs varied significantly. The pre-mixing method, where siRNA and lipids are mixed simultaneously, resulted in a more uniform distribution of siRNA within the LNPs. In contrast, the post-mixing method, where siRNA is added to pre-formed LNPs, led to a heterogeneous distribution with regions of high and low siRNA concentration.
“This heterogeneity can significantly impact the silencing effect of the siRNA. LNPs with a more uniform siRNA distribution are more likely to deliver their therapeutic payload to target cells effectively. This highlights the critical need to optimize preparation conditions for improving therapeutic outcomes,” explains Dr. Ueda.
The findings indicate that pre-mixed LNPs exhibit superior gene-silencing effects. In these LNPs, ionizable lipids were more tightly associated with siRNA, forming a stacked bilayer structure that enhanced gene silencing. In contrast, post-mixed LNPs displayed a more heterogeneous structure, likely impeding their ability to fuse with cell membranes and reducing their therapeutic effectiveness.
“This research could improve people’s lives by enhancing gene therapies and RNA-based medicines. By optimizing how siRNA is delivered using lipid nanoparticles (LNPs), treatments for diseases like cancer, genetic disorders, and viral infections could become more effective. Additionally, it could improve the efficiency and safety of RNA vaccines, like those used for COVID-19, by making them more stable and reducing side effects. Overall, this study has the potential to lead to more effective and safer treatments for patients,” adds Dr. Ueda.
Looking ahead, these advancements could contribute to the development of more personalized medicine, with treatments tailored to individual patients. Enhanced drug delivery systems could also reduce costs and increase access to innovative therapies, benefiting a wider population.
About Assistant Professor Keisuke Ueda
Keisuke Ueda is an Assistant Professor at the Graduate School of Pharmaceutical Sciences, Chiba University. He is engaged in pharmaceutical formulation research, education, and collaborative research with the pharmaceutical industry. He received his Ph.D. in Pharmaceutical Sciences from Chiba University in 2015. He has contributed to more than 90 peer-reviewed publications and serves on the Editorial Advisory Board for Molecular Pharmaceutics (American Chemical Society). In 2021, one of his articles was selected as the ACS Editors’ Choice. In addition, one of his articles in 2022 was featured on the front cover of Volume 19, Issue 1, of Molecular Pharmaceutics.
Funding:
This work was supported by the cSIMVa Vaccine Challenge Grants.
Reference:
Title of original paper: NMR-based analysis of impact of siRNA mixing conditions on internal structure of siRNA-loaded LNP
Authors: Keisuke Ueda, Yui Sakagawa, Tomoki Saito, Fumie Sakuma, Hiroki Tanaka, Hidetaka Akita, Kenjirou Higashi, Kunikazu Moribe
Affiliations:
- Graduate School of Pharmaceutical Sciences, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8675, Japan
- Graduate School of Pharmaceutical Sciences, Tohoku University, 6-3 Aoba Aramaki, Aoba-ku, Sendai City, Miyagi 980-8578, Japan
Journal: Journal of Controlled Release
DOI: 10.1016/j.jconrel.2024.07.055
Contact: Keisuke Ueda
Graduate School of Pharmaceutical Sciences, Chiba University
Email: keisuke@chiba-u.jp
Public Relations Office, Chiba University
Address: 1-33 Yayoi, Inage, Chiba 263-8522 JAPAN
Email: koho-press@chiba-u.jp
Tel: +81-43-290-2018





